Symptoms & Screening
Who Should be Tested?
People with developmental delays, speech delays, seizures, and other undiagnosed symptoms should ask to be screened for a creatine deficiency.
On average, CCDS patients begin to show their first symptom around the age of 1. Unfortunately, most patients don’t receive a CCDS diagnosis until about 3 years of age or later. Early diagnosis of these disorders is essential for prompt initiation of treatment and improved outcomes.
Clinical Symptoms
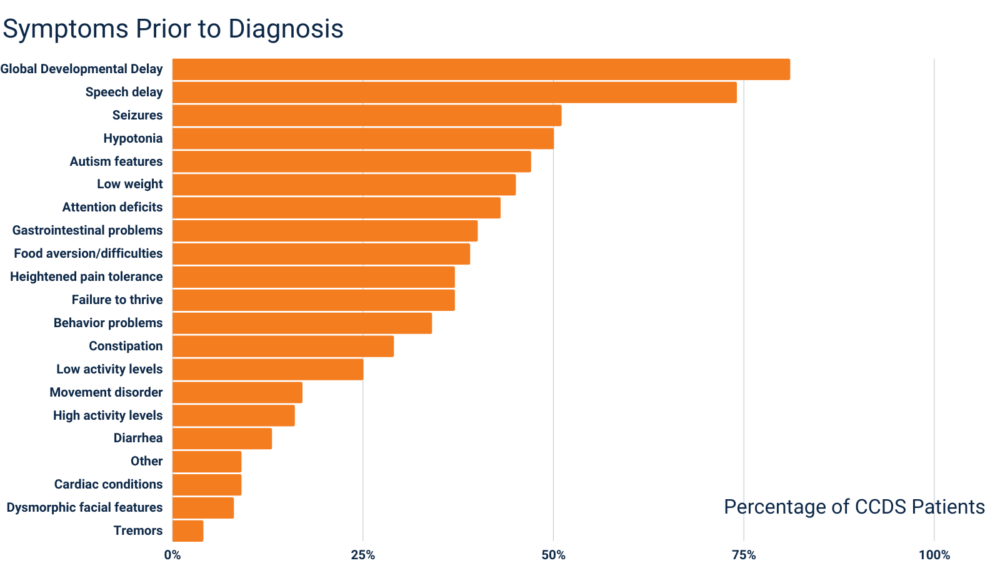
Developmental delay, speech delays, and seizures are the most common symptoms patients experience prior to receiving a CCDS diagnosis. The chart below shows the diverse set of symptoms patients have experienced before receiving a CCDS diagnosis. Please note, there can be variability in the clinical presentation and severity of patients with CCDS.
How to Test for CCDS
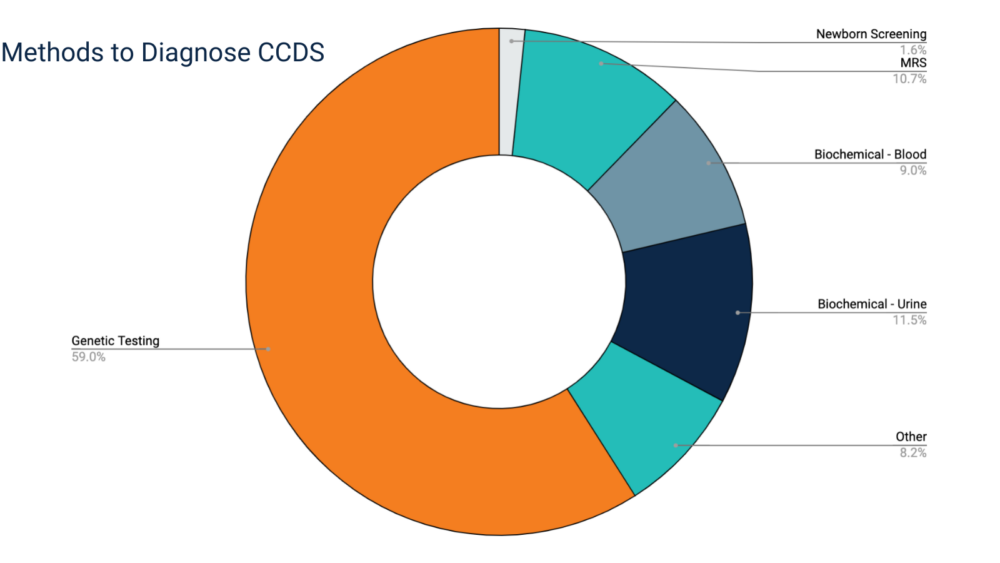
Genetic testing is the most common method used to confirm a CCDS diagnosis. Often, patients will undergo multiple tests that together confirm a CCDS diagnosis (e.g., biochemical testing and an MRS).
Biochemical Testing
Testing in both urine and plasma is recommended to screen for all three disorders. At a minimum, a urine specimen should be tested.
Creatine transporter deficiency (CTD) will be missed if only plasma is screened because males with this disorder have normal creatine in plasma; urine is needed to make this diagnosis in males.
BLOOD PLASMA SCREEN |
||
|---|---|---|
| Guanidinoacetic Acid (GAA or GUAC) | Creatine | |
| Creatine Transporter Deficiency* | Normal | Normal |
| AGAT Deficiency | Low | Low/normal |
| GAMT Deficiency | Elevated | Low |
Urine SCREEN* |
||
|---|---|---|
| Guanidinoacetic Acid (GAA or GUAC) | Creatine | |
| Creatine Transporter Deficiency | Normal | Elevated |
| AGAT Deficiency | Low | Low/normal |
| GAMT Deficiency | Elevated | Low/normal |
*Inclusion of urine screening is recommended because CTD can appear normal in plasma screening. Urine creatine can be normal in females heterozygous for CTD. Sequencing of the SLC6A8 gene is needed for the assessment of females for CTD. Urine metabolites are measured relative to creatinine.
Low plasma and urinary creatinine values are a diagnostic clue for AGAT and GAMT deficiency. Generalized pseudo-elevations of urine metabolites normalized to creatinine, such as amino acids and organic acids, may be observed as a result of low urinary creatinine.
Magnetic Resonance Spectroscopy (MRS)
Decreased or absent creatine peak is noted for all three disorders
Guanidinoacetate peak in GAMT Deficiency


Genetic Testing / Gene Sequencing
Gene sequencing of the following genes may be used to confirm a CCDS diagnosis:
GATM gene > AGAT Deficiency
GAMT gene > GAMT Deficiency
SLC6A8 gene > Creatine Transporter Deficiency (CTD)
Newborn Screening
In 2023, GAMT Deficiency was added to the Recommended Uniform Screening Panel (RUSP), recommending that all babies born in the United States be tested for GAMT as newborns. The RUSP provides state newborn screening programs with a carefully curated list of disorders that meet the committee’s criteria for inclusion. More information can be found here.
Other Methods
Fibroblast assays may be used to measure AGAT and GAMT activity, and to detect creatine uptake for CTD.
Share Your Expertise
The ACD is always looking for experienced clinicians and researchers that share our passion for Cerebral Creatine Deficiency Syndromes. If you would like to be added to our database of CCDS Centers, please submit the form below.
-
Click to Submit Your Information
Submit your information if you are willing to be included in our database of CCDS Centers.
Laboratories
-
Via Biochemical Assessment
Biochemical Assessment for Plasma and Urine GAA/Creatine
- Arup Laboratories – Creatine Disorders Panel (Urine)
- Arup Laboratories – Creatine Disorders Panel (Plasma or Serum)
- Baylor Genetics – Creatine and Guanidinoacitate Test (Urine and Plasma)
- Duke University Health System – Creatine and Guanidinoacetate Test (Urine)
- Duke University Health System – Creatine and Guanidinoacetate Test (Plasma)
- Greenwood Genetic Center
- Kennedy Krieger Institute – Biochemical Testing for Creatine Metabolism Disorders
- Mayo Clinic Laboratories – Creatine Disorders Panel (Plasma)
- Mayo Clinic Laboratories – Creatine Disorders Panel (Urine)
- LabCorp – Plasma Creatine and Guanidinoacetate Test
- LabCorp – Urine Creatine and Guanidinoacetate Test
- MNG Laboratories/ Atlanta Center: Medical Neurogenetics, LLC.,
- Quest Diagnostics – Creatine Biosynthesis Disorders Panel (Urine)
- Quest Diagnostics – Plasma Creatine and Guanidinoacetate Test
- Seattle Children’s Hospital
- University of Alabama Birmingham, Biochemical Genetics Laboratory
-
Via Single Gene Sequencing
Sequencing of GAMT (GAMT Deficiency)
- ARUP Laboratories, Molecular Genetics Laboratory
- Baylor Miraca Genetics Laboratories
- Cincinnati Children’s Hospital Medical Center, Molecular Genetics Laboratory
- Fulgent Diagnostics
- Prevention Genetics
Sequencing of GATM (AGAT Deficiency)
- ARUP Laboratories, Molecular Genetics Laboratory
- Baylor Miraca Genetics Laboratories
- Cincinnati Children’s Hospital Medical Center, Molecular Genetics Laboratory
- Fulgent Diagnostics
- Prevention Genetics
Sequencing of SLC6A8 (Creatine Transporter Deficiency)
- ARUP Laboratories, Molecular Genetics Laboratory
- Baylor Miraca Genetics Laboratories
- Center for Human Genetics, Inc.
- Cincinnati Children’s Hospital Medical Center, Molecular Genetics Laboratory
- Fulgent Diagnostics
- Greenwood Genetic Center Diagnostic Laboratories
*Deletions/Duplications in these genes appear to be extremely rare. However, some of the above laboratories also offer deletion/duplication testing in addition to sequencing, which may need to be ordered separately if desired.
-
Via Multigene Panel
Sequencing Panels that include GAMT
- Ambry Genetics – Comprehensive Epilepsy Testing (EpiNext)
- Courtagen Life Sciences – epiSEEK Focus
- Emory University – Epilepsy and Seizure Disorders Panel
- Emory University – Neurology: Sequencing Panel
- Fulgent Diagnostics – Epilepsy NGS Panel
- Fulgent Diagnostics – Intellectual Disability NGS Panel
- Fulgent Diagnostics – Lysosomal Disorders NGS Panel
- GeneDx – Comprehensive Epilepsy Panel
- GeneDx – Infantile Epilepsy Panel
- Gene Dx – Childhood Onset Epilepsy Panel
- Greenwood Genetic Center Diagnostic Laboratories – NGS Epilepsy/Seizure Panel
- Prevention Genetics – Early Infantile Epileptic Encephalopathy, Recessive NextGen Sequencing (NGS) panel
- Prevention Genetics – Early Infantile Epileptic Encephalopathy NextGen Sequencing (NGS) Panel
Sequencing Panels that include GATM
- Ambry Genetics – Comprehensive Epilepsy Testing (EpiNext)
- Courtagen Life Sciences – epiSEEK Focus
- Courtagen Life Sciences – nucSEEK Comprehensive Sequence Analysis of the Nuclear Mitochondrial Exome
- Emory University – Epilepsy and Seizure Disorders Panel
- Emory University – Neurology: Sequencing Panel
- Fulgent Diagnostics – Epilepsy NGS Panel
- Fulgent Diagnostics – Nuclear-Mito NGS Panel
- GeneDx – Comprehensive Epilepsy Panel
- GeneDx – Childhood Onset Epilepsy Panel
- Greenwood Genetic Center Diagnostic Laboratories – NGS Epilepsy/Seizure Panel
Sequencing Panels that include SLC6A8
- ARUP – X-Linked Intellectual Disability Panel
- Courtagen Life Sciences – devSEEK Sequence Analysis for Neurodevelopmental Disorders, Developmental Delay, Intellectual Disability, Autism Spectrum Disorders
- Courtagen Life Sciences – nucSEEK Comprehensive Sequence Analysis of the Nuclear Mitochondrial Exome
- Fulgent Diagnostics – Intellectual Disability NGS Panel
- Fulgent Diagnostics –Nuclear-Mito NGS Panel
- InVitae – Early Infantile Epileptic Encephalopathy Panel
- Transgenomic – Autism Spectrum Disorder/Intellectual Disability/Multiple Congenital Anomalies NGS Panel
- University of Chicago – Comprehensive Non-Specific ID Sequencing Panel
Sequencing Panels that include all three (GAMT, GATM and SLC6A8)
- Courtagen Life Sciences – epiSEEK Infancy and Childhood Epilepsy Panel
- Courtagen Life Sciences – devACT Clinical Management Panel, Developmental Delay, Intellectual Disability, Autism Spectrum Disorders
- GeneDx – Comprehensive Mitochondrial Nuclear Gene Panel
- GeneDx – Combined Mito Genome Plus Mito Nuclear Gene Panel
- InVitae – Epilepsy Panel
- Transgenomic – NuclearMitome
- Transgenomic – Comprehensive Epilepsy Evaluation NGS Panel
- University of Chicago – Epilepsy Exome Panel
*Whole Exome Sequencing (WES) is also available through many labs, and this test should also detect all three creatine deficiency syndromes.
References
Longo N, Ardon O, Vanzo R, Schwartz E, Pasquali M. Disorders of creatine transport and metabolism. Am J Med Genet C Semin Med Genet. 157C:72-8, 2011.
Mercimek-Mahmutoglu S, Stöckler-Ipsiroglu S, Salomons GS. Creatine Deficiency Syndromes. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2015. 2009 Jan 15.
Rosenberg EH, Almeida LS, Kleefstra T, deGrauw RS, et al. High prevalence of SLC6A8 deficiency in X-linked mental retardation. Am J Hum Genet. 75:97-105, 2004
Stockler-Ipsiroglu S, Apatean D, Battini R, DeBrosse S, et al. Arginine:glycine amidinotransferase (AGAT) deficiency: Clinical features and long term outcomes in 16 patients diagnosed worldwide. Mol Genet Metab. 2015 Oct 17. [Epub ahead of print]
Stockler-Ipsiroglu S, van Karnebeek C, Longo N, Korenke GC, et al. Guanidinoacetate methyltransferase (GAMT) deficiency: outcomes in 48 individuals and recommendations for diagnosis, treatment and monitoring. Mol Genet Metab. 111:16-25, 2014
van de Kamp JM, Betsalel OT, Mercimek-Mahmutoglu S, Abulhoul L, et al. Phenotype and genotype in 101 males with X-linked creatine transporter deficiency. J Med Genet. 50:463-72, 2013
CreatineInfo Patient Registry and Natural History Study (2023).